Staff Introduction
-

-
持田 恵一 Keiichi MOCHIDA
- Emailkeiichi.mochidanagasaki-u.ac.jp- Position / Degree Institute of Integrated Science and Technology, Professor
School of Information and Data Sciences, Professor
Doctor of Science- Specialized Field Genome informatics, Bio-database, Genetics and breeding- External Links researchmap
CV
| Mar.1998 | Yokohama National University, Faculty of Education, Graduated |
| Mar.2000 | Yokohama City University, Graduate School of Integrated Science, Master Course, Completed |
| Mar.2003 | Yokohama City University, Graduate School of Integrated Science, Doctor Course, Completed |
| Mar.2003 | Doctor (Science), Yokohama City University |
| Apr.2003 | University of Tsukuba, Research Associate |
| Apr.2004 | Nagahama Institute of Bio-Science and Technology, Bioinformatics Course, Assistant Professor |
| Oct.2005 | RIKEN, Plant Science Center, Research Associate |
| Oct.2008 | RIKEN, Plant Science Center, Research Scientist |
| Jul.2010 | RIKEN, Biomass Engineering Program, Senior Scientist |
| Apr.2013 | RIKEN, Center for Sustainable Resource Science, Biomass Research Platform Team, Deputy Team Leader |
| Apr.2015 | RIKEN, Center for Sustainable Resource Science, Cellulose Production Research Team, Team Leader |
| Apr.2018 | RIKEN, Center for Sustainable Resource Science, Bioproductivity Informatics Research Team (present) |
| Apr.2021 | Nagasaki University, School of Information and Data Sciences, Professor (present) |
Research Activities
Knowledge discovery from biological deep phenotype data
Comprehensive analyses of various omics spectrums generate diverse and high-dimensional datasets representing physiological states of organisms. Through integrated analysis of multiple omics datasets such as bio-imaging, transcriptome, proteome and metabolome, we aim to identify useful genes and their functions.
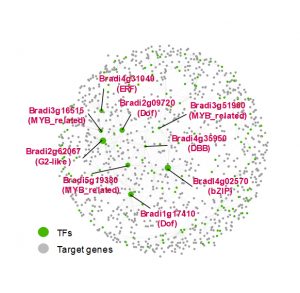
A gene regulatory network estimated based on transcriptome datasets
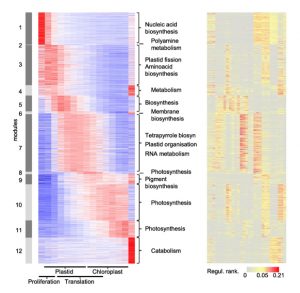
A gene expression map associated with potential transcriptional regulators
Genome-to-phenome modeling in crops
To reveal genetic association between genomic variations and diversities of crop-environment interactions, and diversities of terminal agronomic traits, we aim to computationally modeling genome-to-phenome based on our better understanding of crop-environment interactions along with life-course of crop growth toward crop improvement.
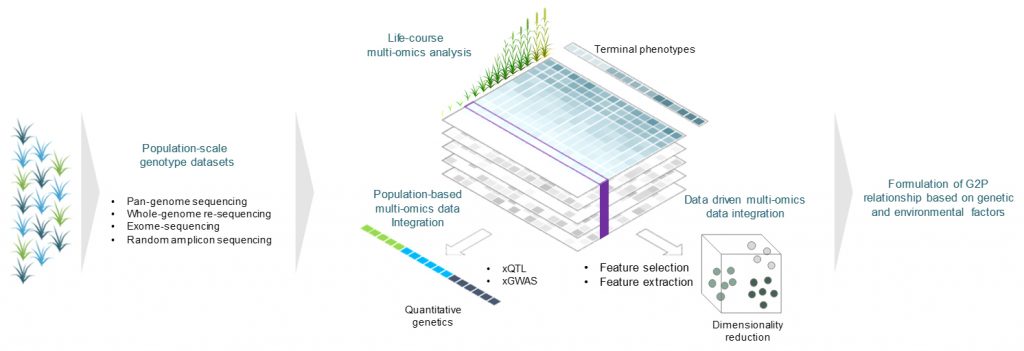
Life-course approach in crops for formulation of genotype-phenotype relationships
Educational Activities
Class
School of Information and Data Sciences:First-year Seminar, Medical and Bio informatics I/Ⅲ, Research Project
